Understanding 23andme Chip Versions: Why V3, V4, V5 and Others Can Differ
Contents
When I first started working on the idea of creating personalized nutrition plans based on consumer genotyping kits I envisaged a relatively straightforward (but long) process. Find the genes and SNPs that we’re interested in, do a load of research about their relative impacts, and then assign them to an algorithm.
So far so good. With this early diet type matrix in hand, we started testing some early kits, and things were going well. There was the odd missing SNP where the test had failed, but other than that, everything was going smoothly, so smoothly that I decided to finally bite the bullet and send off my own sample for testing.
After getting the email I rushed onto 23andme to download my raw data and plugged it straight into the matrix. Where it promptly…failed.
The file reported masses of missing SNPs and made the calculations effectively meaningless. Looking back through the raw data I was able to tell very quickly that I was on a different chip, 23andme v5 to be precise. This sent us back to the drawing board, where we needed to create an algorithm that wouldn’t just work for the select and lucky few who were on v3 or v4 of 23andme, but for people on newer chips, and also other platforms such as ancestry.com.
This is also why we offer test kits at Gene Food for users who want to move beyond just the SNPs listed in raw data from Ancestry and 23andme.
So what is a chip exactly?
Let’s row it back a little bit first and look at what a “chip” actually is.
- When talking about DNA “chip” usually refers to a DNA microarrray, which has been fabricated into a small area, reminiscent of a computer chip.
- Microarrays are made up of grids where each individual cell or spot features thousands of DNA probes that are specific to a particular region of DNA or RNA, in our case we are interested in SNPs.
- When the sample is added to the chip any DNA which matches that of the probes will bind tightly and this will be detected by the machine that reads the chip.
- Now these grids can range in size from a few thousand spots up to several million, with the consumer genetics chips sitting somewhere in the middle at around 500,000 to 1,000,000 spots.
- This is the array aspect of the microarray. As the other part of the name suggests, these are absolutely tiny, often fitting inside a cm2 or smaller.

Comparing 23andme chip versions
So 1,000,000 individual SNPs sounds like a lot right? Well, as we know it’s actually only a fragment of the size of the entire human genome which is thought to weigh in at around 3,300,000,000 (3.3 billion) bases in size, so just a drop in the ocean really.
For this reason, manufacturers often change which SNPs are read on a particular array, or they may custom design one for a company as they require it. There are also several different companies (Illumina and Affymetrix are two of the biggest players) that supply these chips and each will offer different coverage.
So, as a direct to consumer genetic testing company you have a whole host of different options available to you.
Let’s drill it down specifically for 23andme and look at three of their chips, v3, v4 and v5 and see what exactly differs between them.
| Chip Version | Supplier | Chip Name | SNPs | Genefood Coverage |
|---|---|---|---|---|
| v3 | Illumina | OmniExpress+ | 967,000 | 98% |
| v4 | Illumina | OmniExpress+ (Custom) | 570,000 | 95% |
| v5 | Illumina | GSA | 640,000 (+60,000 custom) | 92% |
- With v3 23andme were looking at a huge number of SNPs, so people with that chip come the closest to having 100% coverage in our Gene Food matrix.
- Then in the shift from v3 to v4 we saw a massive cut, reducing the SNP count by nearly half.
- Then with the release of v5 we see an increase in SNP number again. At this point there is also a switch in the platform of choice from Omni Express to the Global Screening Array (GSA).
Why does 23andme keep changing?
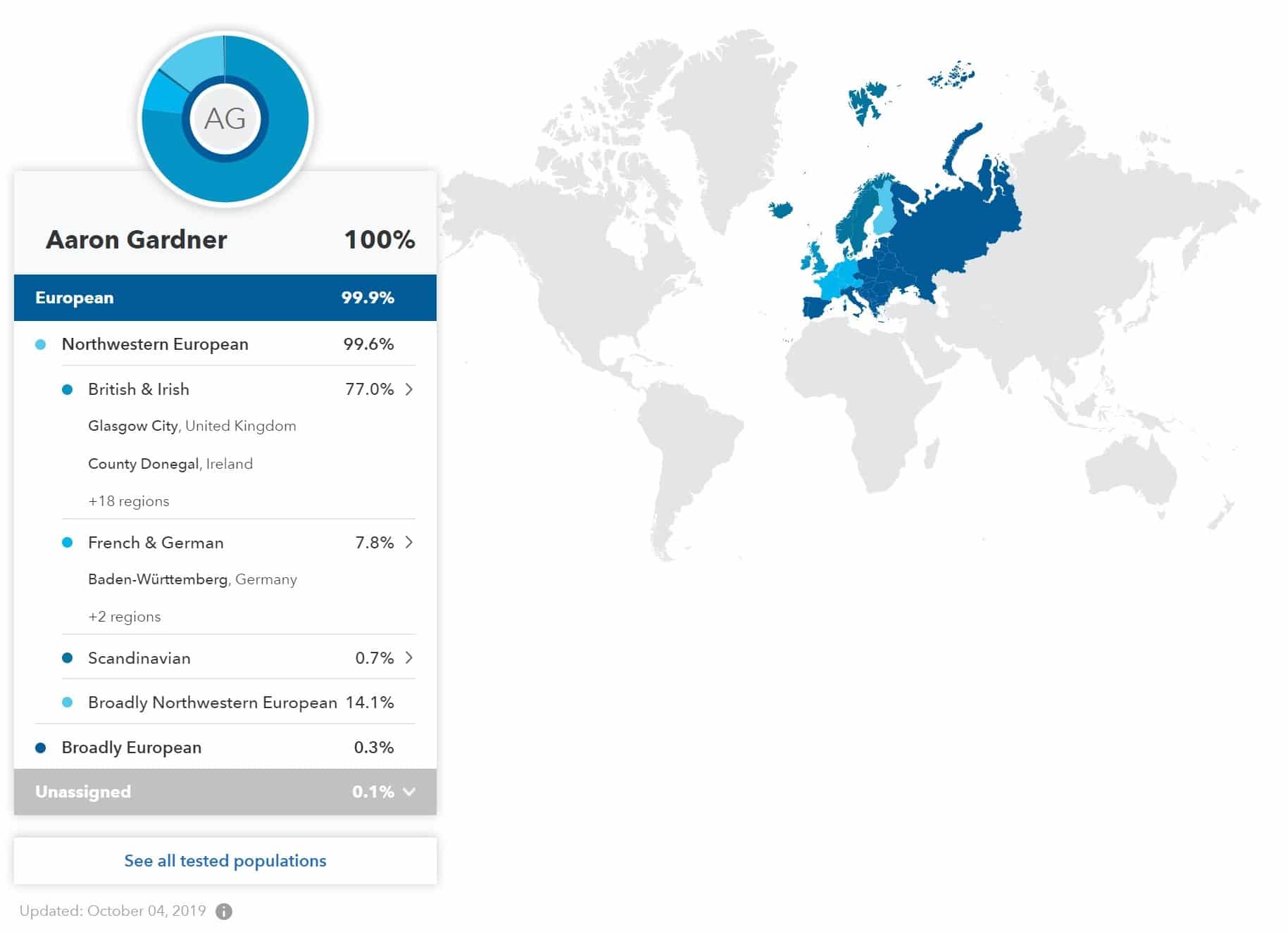
So companies will keep switching things up to try and improve their core product. For 23andme this is a combination of ancestry and geographical tracking and some health reporting. The switch in platforms from Omni Express to GSA is the easiest to explain in this context as the GSA provides much better ancestry information for non-caucasians. If you’ve been a member of 23andme for some time you’ll have seen refinements to your ancestry information during this period and in part this is driven by a shift to a new platform. You can see this newer level of granularity in the ancestry map which now reports regions to a much higher degree. Which can be seen in my (not very exciting) version of the ancestry report. But 23andme’s other angle is health reports, and they are involved in ongoing discussions with the FDA in the US as to what exactly they can provide information about. But in part the shift from v3 to v4 was an attempt to address this and come into compliance with the FDA in 2014.
A large portion of the SNPs cut in the shift from v3 to v4 were health-related SNPs that they are unable to disclose to individuals. Thankfully, these restrictions are being slowly eased as 23andme and more and more health-related tests are appearing on the site again. This is where the more customizable v5 chip comes in, as it allows 23andme to target specific diseases or health effects of interest to them.
Summary
- There are large numbers of DNA chips or microarrays out there, which cover different regions of the genome.
- Suppliers will adjust chips based on regions of DNA and outputs that they’re interested in, such as our array which fits perfectly with our nutrition matrix, but also are coming under greater scrutiny from government bodies such as the FDA as to what they can report.
- This is why versions of 23andme, and other companies like Ancestry, can change from version to version.



